Network model of a bacterial population
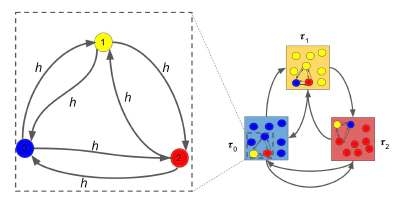
A network model of a bacterial population with phenotypic switching, and some results from this model about the asymtpotic growth rate of the population. This is the code used for this project and a longer description of the project details.
Description
Bacterial populations can be made up of several sub-populations known as phenotypes. The phenotypes are genetically identical, but a given phenotype may possess advantageous traits – such as resistance to antibiotics – that can enable the bacterial population to survive in adverse environments [1]. In the presence of an antibiotic, an individual cell in a bacterial population could switch to a resistant phenotype to maximize the entire population’s chances of survival. We modeled a bacterial population with inter-phenotypic switching as a network, with every node representing a phenotype and every (directed) edge represented the ability of one phenotype to switch to another phenotype. The edge weights encode the rate of switching from one phenotype to another [2]. We observed that as more switching pathways were restricted (which was modeled as removal of edges from the network), the edge weights on the remaining edges would have to be increased, in order to maintain the asymptotic population growth rate.
References
[1] Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006 Jul;4(7):556-62. doi: 10.1038/nrmicro1445. PMID: 16778840.
[2] Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005 Sep 23;309(5743):2075-8. doi: 10.1126/science.1114383. Epub 2005 Aug 25. PMID: 16123265